
In general terms, inbreeding is where the sire and dam have ancestors in common. The level of inbreeding is simply how closely related these relatives are across the sire and dam lines.
The standard (mathematical) measure for the level of inbreeding is the Inbreeding Co-efficient. It indicates the probability (between 0% and 100%) that genes at a randomly chosen location in the DNA are identical by descent. The technique assumes that there are 2 forms of a gene and that each form has an equal chance to be passed on to the next generation.
One limitation on calculating inbreeding coefficients is the depth of pedigrees available. Animals with a shallow pedigree may have a low inbreeding coefficient simply because their related ancestors are not on the database.

Inbreeding Coefficient: is expressed as a percentage value. A low inbreeding coefficient means a low level of inbreeding (eg 3% as in the example above). In most beef cattle breed societies, the vast majority of animals have an inbreeding coefficient of less than 10%, inbreeding coefficients over 30% are unusual, and over 40% are rare.
Generations - Adjacent to the inbreeding coefficient, there are two numbers indicating the minimum number of generations in the animal’s pedigree and, in brackets, the average number of generations in the animal's pedigree - as currently available on the database. Deeper pedigrees will result in a more accurate calculation of the inbreeding coefficient, particularly where the inbreeding coefficient is small.
Typical inbreeding coefficients for various relationships are given in Table 1. The inbreeding coefficient will be higher if the ancestors in the pedigree are also inbred themselves
Table 1. Inbreeding coefficients for various inbred relationships
| Relationship | Inbreeding Coefficient (%) * |
|---|---|
| Animal mated to its own parent (eg Sire / daughter) | 25% |
| Half sib matings (parents have a common sire or dam) | 12.5% |
| Full sib matings (parents have a common sire and dam) | 25% |
| Animal has a single common great grand parent | 3.1% |
* minimum value, will be higher if ancestors are themselves inbred
Not surprisingly, smaller populations tend to have proportionally more animals with higher inbreeding coefficients than larger populations - simply because there are fewer candidate animals to select from (Figure 1).
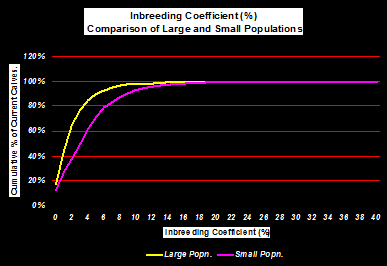
Figure 1. Comparison of Inbreeding Coefficients for large and small population size.
Selection in animal breeding systems uses genetic diversity/variation to improve the population by selecting superior animals for desirable traits. The more genetic variation observed in a population, the bigger the potential genetic gain possible in each generation. Conversely, decreased genetic variation increases the similarities of the population. Inbreeding reduces the amount of genetic diversity in a population.
Explanation of inbreeding in animal populations relies on a few basic genetic principles. Genetic information is stored in Chromosomes. Chromosomes are made up of DNA. Genes are sections of DNA and occur in pairs. A particular gene will occur at a particular site (locus, plural is loci) in the DNA of a particular Chromosome. The different forms of a gene (usually 2) that can occur at that locus are called alleles. Where both alleles are the same at the locus, they are called homozygous. Where the alleles are different, they are called heterozygous. In general, the two alleles will have an equal influence on the performance of an animal. That is, the heterozygous form (both alleles are present) tends to have performance midway between the two homozygous forms. In a few cases, one allele will have the main (dominant) effect on an animal, while the other allele will only have an effect in its homozygous form. These are called dominant and recessive genes.
The coefficient of inbreeding (as proposed by Sewell Wright in 1922) is the probability that two alleles at a randomly chosen locus are identical by descent. Note that alleles may be identical for other reasons, but the inbreeding coefficient is just looking at the mathematical probability that the alleles have come from a common ancestor.
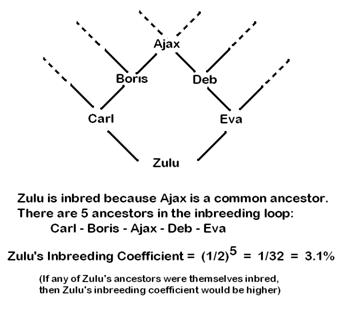
Example of calculating an Inbreeding Coefficient for an animal that has a common great grand parent:
As inbreeding increases, the frequency of alleles being homozygous at a particular locus also increases. Hence inbreeding reduces the amount of variation in a population.
Line breeding attempts to maximise desirable homozygous alleles (ie fix these desirable genes in the population) using selection and planned breeding programs – a sort of controlled inbreeding. However, an increase in homozygous alleles may also increase homozygous alleles with undesirable effects which can result in reduced performance and/or fertility (inbreeding depression); or even be fatal (see lethal recessives).
The converse of inbreeding is out-crossing. Some component of observed hybrid vigour is the out-crossing effect. Note that two inbred animals that are unrelated themselves will have fully out crossed progeny (inbreeding coefficient of zero). Hence, you can go from inbred to out crossed in one generation.
Inbreeding depression is a term used to describe the reduction in performance and viability due to the increase in inbreeding levels (reduced genetic variation). Reproductive fitness tends to be affected more than performance traits in this respect. The extent of inbreeding depression depends on the population being measured, the level of genes already fixed in the population, the frequency of any deleterious genes that may exist in the population and chance effects of which particular genes are affected.
There is no defined limit as to what is an acceptable level of inbreeding in domestic animal populations. However, inbreeding depression is likely to be more apparent once inbreeding levels get to above 10%. As a very rough guide, there is often a 2-20% decrease in performance of the trait per 10% of inbreeding coefficient.
Some undesirable recessive genes are only expressed in their homozygous form. In these cases, the dominant, desirable allele of the gene is expressed in the animal for both the dominant homozygous and heterozygous forms.
The Arthrogryposis Multiplex (AM) gene is an example of a recessive gene that is lethal. The undesirable allele (a) in its homozygous form (aa) causes the affected animal to have a bent and twisted spine and limited muscle development, but with apparent normal brain tissue. Affected calves are generally still born. Conversely, animals with at least one copy of the dominant allele (A) are normal, unaffected calves (ie the homozygous AA or heterozygous Aa forms of the gene). The heterozygous animals (Aa) are called carriers as they have the potential to pass on the undesirable form (a) of the gene undetected to the next generation.
A carrier (Aa) animal mated to a homozygous normal (AA) animal will result in a 50% chance of the progeny being homozygous normal (AA) and 50% chance of being a heterozygous carrier (Aa) with no lethal recessive (aa) forms (Table 2).. However, a carrier (Aa) mated to another carrier (Aa) has a 25% chance of producing homozygous normal (AA) offspring, 50% chance of producing carrier (Aa) offspring and 25% chance of producing affected (aa) offspring - which die at birth
Table 2. Probability of mating outcomes of heterozygous carriers and homozygous non-carriers
| Mating | Non-carrier (AA) | Carrier (Aa) | |||
|---|---|---|---|---|---|
| outcomes | A | A | A | a | |
| Carrier | A | AA | AA | AA | Aa |
| (Aa) | a | Aa | Aa | Aa | aa |
| Outcomes: | AA 50%, Aa 50% | AA 25%, Aa 50%, aa 25% | |||
A genetic test is now available to detect which alleles the animal actually has - ie whether an animal is homozygous for the desirable allele (AA) or is a carrier (Aa).